CHPC - Research Computing and Data Support for the University
In addition to deploying and operating high performance computational resources and providing advanced user support and training, CHPC serves as an expert team to broadly support the increasingly diverse research computing and data needs on campus. These needs include support for big data, big data movement, data analytics, security, virtual machines, Windows science application servers, protected environments for data mining and analysis of protected health information, and advanced networking.
If you are new to CHPC, the best place to start to get more information on CHPC resources and policies is our Getting Started page.
Upcoming Events:
CHPC Downtime: Tuesday March 5 starting at 7:30am
Posted February 8th, 2024
Two upcoming security related changes
Posted February 6th, 2024
Allocation Requests for Spring 2024 are Due March 1st, 2024
Posted February 1st, 2024
CHPC ANNOUNCEMENT: Change in top level home directory permission settings
Posted December 14th, 2023
CHPC Spring 2024 Presentation Schedule Now Available
CHPC PE DOWNTIME: Partial Protected Environment Downtime -- Oct 24-25, 2023
Posted October 18th, 2023
CHPC INFORMATION: MATLAB and Ansys updates
Posted September 22, 2023
CHPC SECURITY REMINDER
Posted September 8th, 2023
CHPC is reaching out to remind our users of their responsibility to understand what the software being used is doing, especially software that you download, install, or compile yourself. Read More...News History...
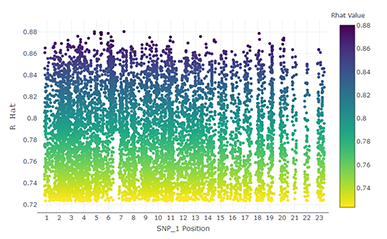
Using Computation to Sort Through Billions of Single-Nucleotide Polymorphism Pairs Rapidly
By Randall Reese1,2, Xiaotian Dai2, and Guifang Fu2
1Idaho National Laboratory, Department of Mathematics and Statistics, 2Utah State University
The interaction between two statistical features can play a pivotal role in contributing to the variation of the response, yet the computational feasibility of screening for interactions often acts as an insurmountable barrier in practice. We developed a new interaction screening procedure which is significantly more tractable computationally. Using the supercomputing resources of the CHPC, we were able to apply our method to a data set containing approximately 11 billion pairs of single nucleotide polymorphisms (SNPs). The goal of this analysis was to ascertain which pairs of SNPs were most strongly associated with the likelihood of a human female developing polycystic ovary syndrome. This distributed computing process allowed us to select in just over two days a set of around 10,000 SNP interaction pairs which factor most strongly into the response. What may have previously taken several weeks (or months) to obtain now took less than three days. We concluded our analysis by using an implementation of multi-factor dimension reduction (MDR) on the previously aforementioned results.
System Status
General Environment
| General Nodes | ||
|---|---|---|
| system | cores | % util. |
| kingspeak | 698/972 | 71.81% |
| notchpeak | 2922/3212 | 90.97% |
| lonepeak | 3088/3140 | 98.34% |
| Owner/Restricted Nodes | ||
| system | cores | % util. |
| ash | 1152/1152 | 100% |
| notchpeak | 17847/18328 | 97.38% |
| kingspeak | 1429/5340 | 26.76% |
| lonepeak | 0/416 | 0% |
Protected Environment
| General Nodes | ||
|---|---|---|
| system | cores | % util. |
| redwood | 41/616 | 6.66% |
| Owner/Restricted Nodes | ||
| system | cores | % util. |
| redwood | 1342/6280 | 21.37% |