Molecular Level Study of Epitope Mimicry Leading to Onset of Type 1 Diabetes
Research by Ryan Gardner1, Joshua Wilkins2, Sejal Mistry3, Ram Gouripeddi3,4, and Julio C. Facelli3,4
1Weber State University, 2North Carolina A&T, 3University of Utah Department of Biomedical Informatics, 4Utah Clinical and Translational Science Institute
Ryan Gardner, Joshua Wilkins, Sejal Mistry, Ram Gouripeddi, and Julio C. Facelli studied 35 potential molecular mimics that exhibited sequence homology. The team calculated their structures, electrostatic properties, and hydrophobicity to gain a better understanding of their characteristics and relation to the onset of Type 1 Diabetes.
Gardner et al. used the I-TASSER suite to calculate the structures of the epitope polypeptides. The structures were analyzed with ChimeraX to superimpose each protein pair by creating a pairwise sequence alignment, then fitting the aligned residue pairs. Once the available matches were made, the researchers calculated the root mean square deviation (RMSD) of each pair. Finally, they calculated the electrostatic and hydrophobic properties of each epitope using ChimeraX.
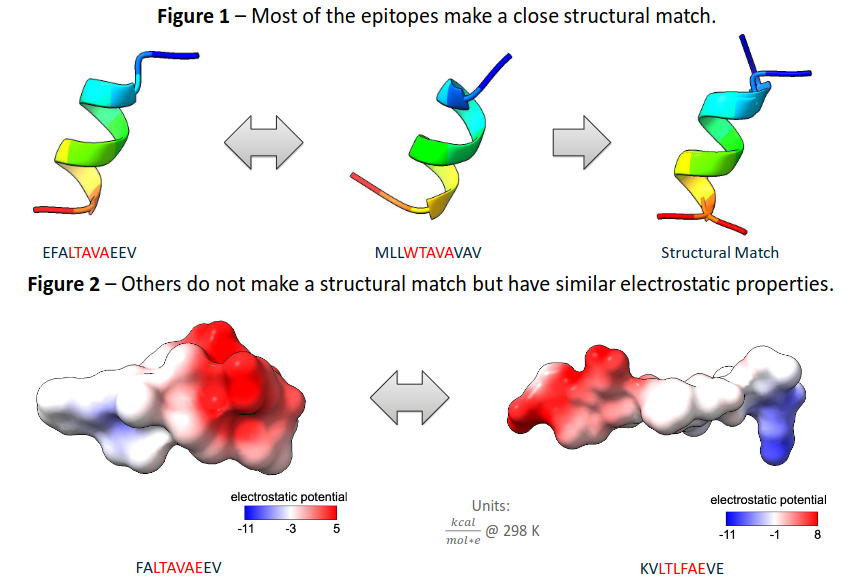
The homology pipeline of Mistry et al. has proven successful for 83% of epitope pairs, yielding a close structural match with an RMSD < 1.5 Å.
Despite remaining structurally unmatched, the 6 pairs that retained similar electrostatic and hydrophobic values imply that they may still successfully bind to MHC molecules.
As a future direction, Gardner et al. plan to cross-check their results with AlphaFold to ensure the reliability of the data obtained. Should AlphaFold yield divergent results, it emphasizes the importance of further investigation, especially regarding the docking process.
Attribution: This content was provided by researchers involved with the project and edited by staff at the CHPC.
